

Gurliaccio Main Page | FamilyTree Italy DNA Project
My Gurliaccio Y DNA
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Advanced Y DNA Markers
| ||||||||||||||||||||
| ||||||||||||||||||||
|
My Haplogroup
|
||||||||||||||||||||
|
J |
The age of this haplogroup is approx. 20,000 years old and can be split into two main subgroups, J1 and J2 which are thought to have origins in the southern and northern parts of the Fertile Crescent respectively. DNA Haplogroup J is found at highest frequencies in Middle Eastern and north African populations where it most likely evolved. This marker has been carried by Middle Eastern traders into Europe, central Asia, India, and Pakistan. |
|
J2 |
According to DNA this lineage originated in the northern portion of the Fertile Crescent where it later spread throughout central Asia, the Mediterranean, and south into India. J2 has a high frequency across Iberia, Italy, Turkey, Albania, Greece, Arabic countries and across into India. |
|
|
|
Eupedia says this. For the complete article go here. 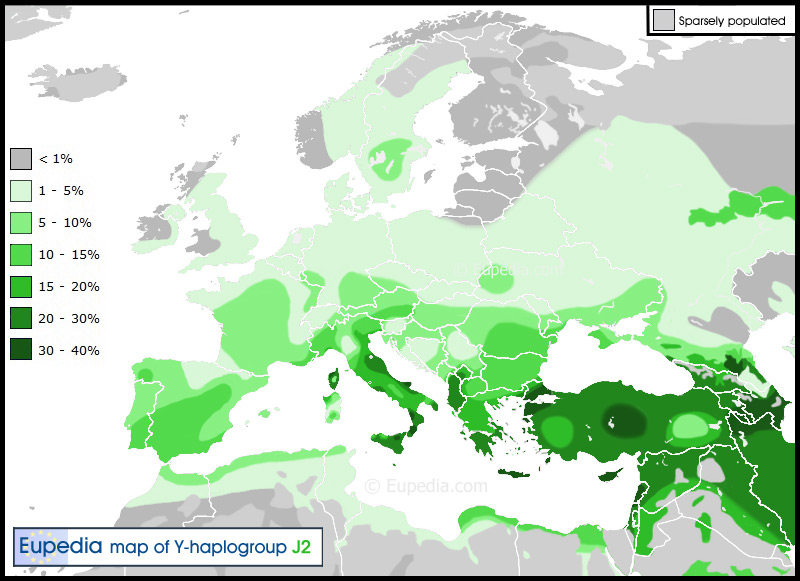
The geographic distribution of J2a bears a strong correlation with the diffusion of agriculture from northern Mesopotamia (where it peaks) towards Anatolia, Greece, the whole Middle East, Iran, Afghanistan, Pakistan and western India. Its strong presence in Italy is owed to the migration of the Etruscans from the Near East to central and northern Italy, and to the Greek colonisation of southern Italy. The Phoenicians, Jews, Greeks and Romans all contributed to the presence of J2a in Iberia. The particularly strong frequency of J2a and other Near Eastern haplogroups (J1, E1b1b, T) in the south of the Iberian peninsula, suggest that the Phoenicians played a more decisive role than other peoples. This makes sense considering that the Phoenicians/Carthaginians were the first to arrive, founded the greatest number of cities (including Gadir/Cadiz, Iberia's oldest city), and their settlements match almost exactly the higher frequency zone of southern Analusia. The Romans surely helped spread haplogroup J2 within their borders, judging from the distribution of J2 within Europe (frequency over 5%), which bears an uncanny resemblance to the borders of the Roman Empire. The world's maximum concentrations of J2a is in Crete (32% of the population). The subclade J2a3d (M319) appears to be native to Crete. J2a also reaches high frequencies in Anatolia and the southern Caucasus. Haplogroup J with L24+ clade The highest L24(M530) frequency observed, anywhere in the world, was for Zoroastrians in the city of Yazd, Iran. The frequency was 17.5% out of a random sample of 80. The Persians of Yazd also showed an L24(M530) frequency of 17.5%. Zoroastrians of Tehran showed an L24(M530) frequency of 15.4%. Other areas of Iran showed high frequencies as well, higher than found anywhere else in the extensive set of countries studied in the paper. It seems clear that Zoroastrians and Persians are important groups in our ancient L24(M530) paternal ancestry. Iran then is our most ancient paternal homeland, but can one point to a specific area within Iran as the origin of L24(M530)? Yes, it is possible from the variance. The place of highest variance, highest diversity of haplotypes, is likely the place of origin of L24(M530). Our L24(M530) origin appears to be with the people occupying the Mazandaran Province of Iran where the variance was found to be 0.730. Indigenous peoples include the Mazanderanis who occupy the southern coastal plains of the Caspian Sea and the adjacent Alborz mountains. A closely related ethnic group are the Gilakis. The Gilaki are found in Mazandaran province and also in the Gilan Province just west of Mazandaran Province. These people are the oldest members of L24(M530) and I wonder how many will be found to carry the L25 SNP which is immediately downstream of L24(M530). So far in our FTDNA J-L24-Y-DNA Project we have found only two confirmed haplotypes who are L24+ and not also L25+. The second highest variance (0.651) was found among Persians/Zoroastrians of Yazd and Fars Provinces. The Mazanderanis people and the Persians are no doubt paternally related. |